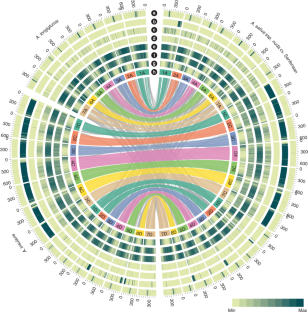
Assemblies of hexaploid cultivated oat, and of close relatives of its diploid and tetraploid progenitors, have revealed its polyploid formation and subgenome evolution. These high-quality oat reference genomes will facilitate the discovery of candidate genes that underlie beneficial traits such as hulless grain and disease resistance.
This is a preview of subscription content
Access options
Subscribe to Journal
Get full journal access for 1 year
59,00 €
only 4,92 € per issue
Tax calculation will be finalised during checkout.
Buy article
Get time limited or full article access on ReadCube.
$32.00
All prices are NET prices.
References
-
Van de Peer, Y. et al. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017). This Review correlates polyploidization with environmental change or stress and its short-term adaptive potential.
-
Yu, H. et al. A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156–1170 (2021). This paper reports a strategy for the polyploidization of diploid rice that can be developed into a staple cereal to strengthen world food security.
-
Appels, R. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018). This is a report of an annotated reference sequence representing the hexaploid bread wheat genome, which improved the understanding of wheat biology and genomics-assisted breeding.
-
Chaffin, A. S. et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 9, 2 (2016). This paper reports the rearrangement of the hexaploid oat genome relative to its ancestral diploid genomes as a result of frequent translocations among chromosomes.
-
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020). This paper reports the telomere-to-telomere assembly of a human chromosome, which was enabled by ultra-long-read nanopore sequencing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Peng, Y. et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. https://doi.org/10.1038/s41588-022-01127-7 (2021).
Rights and permissions
About this article
Cite this article
Genome origin and evolution of common oat. Nat Genet (2022). https://ift.tt/vHY1Moc
-
Published:
-
DOI: https://ift.tt/vHY1Moc
"oat" - Google News
July 25, 2022 at 10:52PM
https://ift.tt/akHX59M
Genome origin and evolution of common oat - Nature.com
"oat" - Google News
https://ift.tt/CylXmSB
https://ift.tt/gD5PjfY
Bagikan Berita Ini














0 Response to "Genome origin and evolution of common oat - Nature.com"
Post a Comment